Undergraduate Spotlight
Julia Phelan
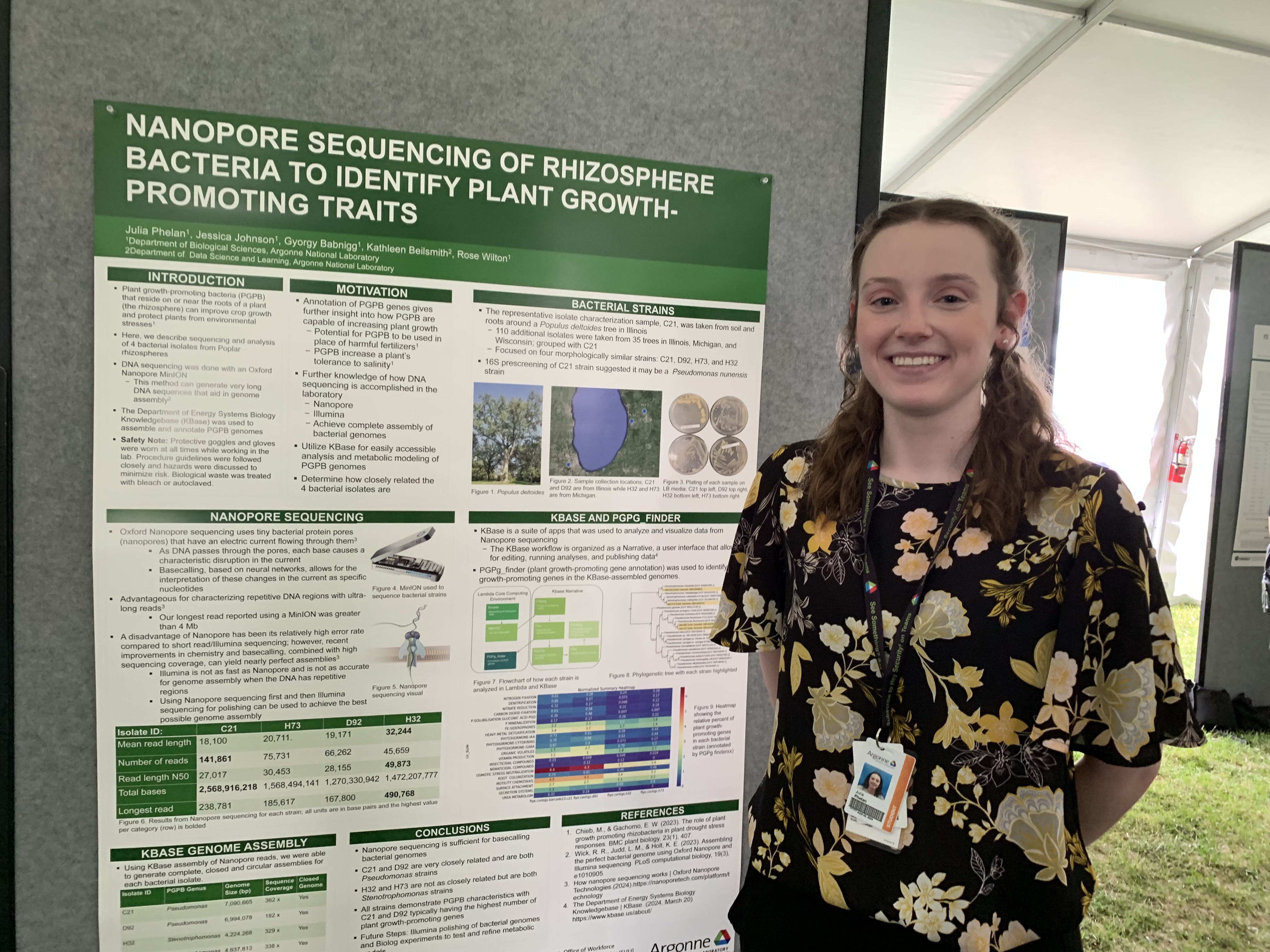
Julia Phelan had an internship at Argonne National Lab this summer. Julia was able to determine the relatedness of four uncharacterized bacterial isolates, as well as confirm that they are plant growth-promoting bacteria, by using Oxford Nanopore sequencing. Analysis of the data was done using KBase, the Department of Energy's Systems Biology Knowledgebase. Here is a link to the published KBase narrative (account required).

Plant growth-promoting bacteria (PGPB) in the rhizosphere (root zone) of plants have varying traits that increase plant growth. These bacteria have the potential to replace harmful fertilizers and protect plants from environmental stresses. However, the mechanisms that PGPB employ are not fully understood and thus pose a challenge for implementation in agricultural settings. Therefore, DNA sequencing of PGPB is necessary for identifying plant growth-promoting traits. This work uses Oxford Nanopore sequencing of four morphologically similar bacterial isolates collected from Poplar tree rhizospheres. The Department of Energy Systems Biology Knowledgebase (KBase) was then used to assemble the bacterial genomes, analyze and visualize the data from Nanopore sequencing. Complete, closed circular genome assemblies were obtained for each isolate. Lastly, the assembled genomes were run through PGPg_finder for a final analysis of the data. PGPg_finder annotation of bacterial genomes allows one to distinguish between each bacterial isolate through the amount and types of plant growth-promoting genes present.